We are looking for a motivated and ambitious postdoctoral researcher to help us in a highly interdisciplinary project. If you are a chemist with experience in microfluidics, lab-on-a-chip systems, or flow chemistry, this position may be for you. An interest in spectroscopy and magnetic resonance is a plus, but prior experience in these fields is not a requirement. More details can be found here.
Author: Marcel
Quantum Mechanics
Lectures on Quantum Mechanics
Quantum Mechanics I
Marcel is teaching quantum mechanics to Chemistry students at various levels. Quantum mechanics forms the theoretical basis for much of chemistry, reaction kinetics, statistical thermodynamics, and spectroscopy. The emphasis on all teaching activities in this field is on understanding the fundamental concepts. This can be a challenge, since the important intellectual steps are easily obscured by the need to resort to rather advanced mathematical formalism. Marcel tries to carefully balance this, without oversimplifying the language.
Lecture notes on introductory quantum mechanics
Quantum Mechanics III – Video Tutorials
These tutorials have been created in the context of the 3-rd year lecture course “Atoms, Spins, and Molecules”, co-taught in the spring semester by Prof. Malcolm Levitt and Marcel Utz.
PhD Position: Hyperpolarisation on a Chip
This position has been filled and is no longer available.
The central aim of this project is to enable hyperpolarised NMR spectroscopy in microfluidic lab-on-a-chip devices.
Nuclear magnetic resonance (NMR) is one of the most versatile tools known to science. It is non-invasive, allowing direct and quantitative studies of metabolic processes and transport phenomena in live systems. We have recently demonstrated thisat small scale, in the context of microfluidic lab-on-a-chip devices. However, such small-scale applications are limited by the comparatively low sensitivity of NMR. Many important processes in living systems happen on a time scale of minutes to hours. They are difficult to probe with NMR, since the nuclear spin states tend to relax to thermal equilibrium within a few seconds.

The aim of this PhD project is to develop an integrated microfluidic lab-on-a-chip platform for high-resolution nuclear magnetic resonance (NMR) spectroscopy and imaging. This will enable direct NMR investigation of miniaturised lab-on-a-chip culture devices for cells, cell aggregates, and tissues. Such devices will find important applications in the life sciences, for example as disease models, supporting drug discovery and safety testing, as well as tissue engineering.
Hyperpolarisation of the nuclear spins can provide up to 5 orders of magnitude of signal enhancement. However, existing hyperpolarisation techniques are not easily combined with microfluidic systems. The present project will address this by developing a microfluidic device that integrates the chemical reactions and the spin manipulations required for para-hydrogen induced hyper polarisation (PHIP). We intend to combine this with recent developments of molecules that support very long-lived nuclear spin states in order to explore transport and chemical processes in microfluidic devices.
The magnetic resonance research section at the University of Southampton, UK, has access to a wide variety of state-of-the-art magnetic resonance instruments including NMR and EPR spectrometers, as well as hyperpolarization technologies such as dynamic nuclear polarization (DNP) and parahydrogen-induced polarization (PHIP). In addition, Southampton provides an ideal environment for microfluidic technology, with several internationally renowned research groups active in this field.
A 3-year PhD studentship is available in the group of Marcel Utz, supported in part Bruker UK Ltd. The successful candidate should have a good degree in chemistry, or a related field, and a special interest in nuclear magnetic resonance spectroscopy and/or microfluidic systems. The project welcomes applicants from the EU/UK who have or expect to obtain a first class degree in Chemistry, Physics or allied subjects/relevant disciplines. Funding will cover fees and a living allowance in line with RCUK guidelines (£14553 for 2017/2018).
Applications for a PhD in Chemistry should be submitted online at https://studentrecords.soton.ac.uk/BNNRPROD/bzsksrch.P_Search
Please ensure you select the academic session 2018-2019 in the academic year field and click on the Research radio button. Enter Chemistry in the search text field.
General enquiries should be made to Marcel Utz at marcel.utz@soton.ac.uk. Any queries on the application process should be made to pgafnes@soton.ac.uk
Applications will be considered in the order that they are received, and the position will be considered filled when a suitable candidate has been identified.
PhD Position: Protein NMR On A Chip
This position has been filled and is no longer available.
A 3-year PhD studentship is available in the group of Marcel Utz, supported by the Institute for Life Sciences (IfLS). The successful candidate should have a good degree in chemistry, biochemistry, physical chemistry, or biophysics and a special interest in nuclear magnetic resonance spectroscopy and/or microfluidic systems.
 Nuclear magnetic resonance (NMR) spectroscopy is one of the most powerful analytical tools available to study the dynamics, structure, and interactions of proteins. In addition to detailed 3D structural information, NMR is particularly apt at providing information on dynamic processes, as well as binding interactions with peptides and other substrates. NMR is inherently non-invasive. In sharp contrast to other techniques such as x-ray diffraction, which require crystalline samples, high vacuum, and employ intense ionising radiation, NMR allows the study of proteins (and other systems) under very mild and realistic conditions, in some cases even directly in-vivo. The main aim of the present project is to bring this advantage to bear in the context of microfluidic lab-on-a chip devices.
Nuclear magnetic resonance (NMR) spectroscopy is one of the most powerful analytical tools available to study the dynamics, structure, and interactions of proteins. In addition to detailed 3D structural information, NMR is particularly apt at providing information on dynamic processes, as well as binding interactions with peptides and other substrates. NMR is inherently non-invasive. In sharp contrast to other techniques such as x-ray diffraction, which require crystalline samples, high vacuum, and employ intense ionising radiation, NMR allows the study of proteins (and other systems) under very mild and realistic conditions, in some cases even directly in-vivo. The main aim of the present project is to bring this advantage to bear in the context of microfluidic lab-on-a chip devices.
 The ability to perform meaningful NMR studies of protein systems on a chip would open interesting possibilities. For example, binding studies of a protein with an entire library of peptides could be efficiently integrated on a chip, offering considerable savings of cost and time.
The ability to perform meaningful NMR studies of protein systems on a chip would open interesting possibilities. For example, binding studies of a protein with an entire library of peptides could be efficiently integrated on a chip, offering considerable savings of cost and time.
Protein NMR almost universally relies on isotopically labelled samples. Most commonly, proteins are expressed in E. coli cultures fed with universally 15N and 13C labelled amino acids. This process is costly, and the ability to work with very small samples is therefore inherently attractive. Moreover, many proteins of interest are difficult to express in large quantities.
The present project is designed to explore quantitatively the technical challenges that need to be overcome to make microfluidic protein NMR possible. With the specific aim of carrying out a repeated dilution protein binding study, it is designed

to produce a convincing demonstration of the potential of this approach.
The magnetic resonance research section at the University of Southampton, UK, has access to a wide variety of state-of-the-art magnetic resonance instruments including NMR and EPR spectrometers, as well as hyperpolarization technologies such as dynamic nuclear polarization (DNP) and parahydrogen-induced polarization (PHIP). In addition, Southampton provides an ideal environment for microfluidic technology, with several internationally renowned research groups active in this field.
The project welcomes applicants from the EU/UK who have or expect to obtain a first class degree in Chemistry, Physics or allied subjects/relevant disciplines. Funding will cover fees and a living allowance in line with RCUK guidelines (£14553 for 2017/2018).
Applications for a PhD in Chemistry should be submitted online at https://studentrecords.soton.ac.uk
Please ensure you select the academic session 2018-2019 in the academic year field and click on the Research radio button. Enter Chemistry in the search text field.
Spin simulations in Julia
Julia is a programming language designed for numerical and scientific computation. It combined the convenience of an interpreted scripting language with the efficiency of compiled ones like C, Fortran, or C++. The goal of this effort is to provide a very simple and lightweight infrastructure for quick, but accurate, quantum mechanical simulations of small spin systems. NMR.jl represents spin operators in a Hilbert basis formed from the Kronecker
products of the individual \(|\alpha\rangle, |\beta\rangle,\dots\) spin states. It uses sparse matrices wherever possible. In this way, simulations of up to ten spins 1/2 tend to run in a matter of minutes. Larger spin systems, up to about twenty spins or so, are possible, but require significant computational resources.
Here is an example of such a calculation. These commands are meant to be run either as a Julia script, line-by-line on the Julia command line, or, most usefully, as an IJulia notebook in a web browser. If you have not done so, install the current version of NMR.jl in your Julia environment by running the Julia command
Pkg.clone("https://github.com/marcel-utz/NMR.jl");
This will take a few moments. Once it’s done, you are ready to go.
First, we create a clean workspace and declare usage of the NMR modules SpinSim and PauliMatrix:
workspace(); using NMR.SpinSim; using NMR.PauliMatrix;
Then, we need to define the properties of the spin system. In the present case, we are modelling hydroxy-ethyl propionate (HEP). The molecule looks like this:
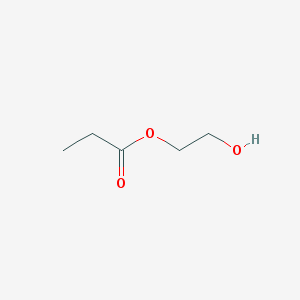
There are five protons on the propionate side, three methyl (a) and two methylene (b), and two pairs of methylene protons on the ethyl (right) side (c) and (d). We assume the hydroxyl proton to be in fast exchange with the solvent. In addition, there is a 13C label at the carboxylate site (x). Therefore, we are simulating a ten spin system overall.
The parameters are
Jaa=-12.4; Jab=7.5; Jax=7.2; Jbb=-10.8; Jbx=-5.6; Jcc=-10.8; Jcd=7.5; Jcx=3.2; Jdd=-12.4; Jdx=1.7; δa=1.25; δb=4.15; δc=2.3; δd=1.8;
The Zeeman Hamiltonian (in the rotating frame) is defined by taking into account the chemical shifts of the protons and the Z component of the spin operators:
Hz=δa*(SpinOp(10,Sz,1)+SpinOp(10,Sz,2)+SpinOp(10,Sz,3))+ δb*(SpinOp(10,Sz,4)+SpinOp(10,Sz,5))+ δc*(SpinOp(10,Sz,7)+SpinOp(10,Sz,8))+ δd*(SpinOp(10,Sz,9)+SpinOp(10,Sz,10)) ;
In addition, we need the J coupling Hamiltonian. We use strong coupling between chemically equivalent protons, and weak coupling in all other cases:
HJHHstrong= 2*pi*Jaa*(OpJstrong(10,1,2)+OpJstrong(10,2,3)+OpJstrong(10,1,3))+ 2*pi*Jbb*OpJstrong(10,4,5)+ 2*pi*Jcc*OpJstrong(10,7,8)+ 2*pi*Jdd*OpJstrong(10,9,10); HJHHweak= 2*pi*Jab*sum([OpJweak(10,k,l) for k=1:3,l=4:5])+ 2*pi*Jcd*sum([OpJweak(10,k,l) for k=7:8,l=9:10]);
We also need some of the total spin operators, such as \(F_z\) and \(F_y\):
FzH=sum([SpinOp(10,Sz,k) for k=1:5])+sum([SpinOp(10,Sz,k) for k=7:10]) FyH=sum([SpinOp(10,Sy,k) for k=1:5])+sum([SpinOp(10,Sy,k) for k=7:10]) FmH=sum([SpinOp(10,Sm,k) for k=1:5])+sum([SpinOp(10,Sm,k) for k=7:10]) Fz=sum([SpinOp(10,Sz,k) for k=1:10])
Here, FzH is the \(F_z\) operator for the protons only, where as Fz applies
to all spins, including 13C.
Overview
The Utz lab is a research group within the School of Chemistry at the University of Southampton. It is one of five research groups that together make up the magnetic resonance research section at the University (MagRes@Soton).
 Our research mainly concerns the study of live systems (cells, tissues, and small animals) by nuclear magnetic resonance. We are developing miniaturised “Lab-on-a-chip” devices for the culture of such biological systems, and integrate them with in-situ observation by nuclear magnetic resonance. This combination of microfluidic culture technology with advanced nuclear magnetic resonance techniques provides unique insight into the metabolic processes of life. Our work is motivated by applications across the life sciences, in particular in drug discovery and safety testing, and in the development of disease models.
Our research mainly concerns the study of live systems (cells, tissues, and small animals) by nuclear magnetic resonance. We are developing miniaturised “Lab-on-a-chip” devices for the culture of such biological systems, and integrate them with in-situ observation by nuclear magnetic resonance. This combination of microfluidic culture technology with advanced nuclear magnetic resonance techniques provides unique insight into the metabolic processes of life. Our work is motivated by applications across the life sciences, in particular in drug discovery and safety testing, and in the development of disease models.
Other research areas include the study of complex materials such as hydrogels, solid polymers, and composites. We are also actively contributing to an effort to develop a novel approach to neurostimulation of the spinal cord.
Billy Hale
Billy Hale joined the research group as a postgraduate (PhD) student in 2015, after having earned an MChem degree from the University of Southampton. His research is focused on studying live systems in microfluidic perfusion by NMR.
Ali Yilmaz
Ali obtained his PhD from the University of Copenhagen in 2012, working on NMR metabolomic observations. After a first postdoc at SUNY Buffalo, he joined the Utz research team at Southampton from 2014 until 2016. His main research interests include microfluidic systems, metabolomics, and micro-fabrication techniques for biocompatible microdevices.
Marcel Utz
Vita
Marcel obtained a MSc in materials science from ETH Zürich in 1994. He then joined the research groups of Prof. Ueli Suter in Polymer Science, and of Prof. Richard R. Ernst in Physical Chemistry at ETH as a doctoral candidate. In 1998, he obtained his doctorate, with a thesis on solid-state NMR investigations of glassy polymers under plastic deformation. In 1999, he joined the group of Prof. Pablo DeBenedetti at Princeton University as a postdoc, with a stipend from the Swiss National Science Foundation. A year later, he joined the Institute of Materials Science and the Department of Physics at the University of Connecticut as an Assistant Professor. He was promoted to Associate Professor with Tenure in 2006. In the same year, he moved to the University of Virginia, where he joined the department of Mechanical and Aerospace Engineering. In 2012, Marcel moved to the UK, joining the School of Chemistry at the University of Southampton. He was promoted to a personal chair in 2014. He currently heads the section of Magnetic Resonance within the School of Chemistry. Marcel lives in Winchester, UK, with his wife Song and their son KangKang.
Research Interests
Our main focus lies on the integration of microfluidic lab-on-a-chip technology with NMR spectroscopy and imaging. We believe that there is great potential for NMR in the context of LoC devices, due to its versatility, resolution, and non-invasive nature, which make it an ideal tool to study complex living systems.
In addition, we are also interested in the behaviour of complex materials. Recent examples of this include polyelectrolyte hydrogels and brick-and-mortar type micro-composites.
Welcome
to the Utz research group’s blog site. We post recent advances, insights, and news about our activities here. Thank you for visiting!

